Big tree tutorial¶
This tutorial discusses how to load data from a large tree efficiently using TopiaryExplorer and QIIME. This process requires filtering the tree to include fewer tips that are representative of the full diversity of the tree. The figures used in this tutorial are derived from applying this process to the full Greengenes tree (408,135 tips).
Assumptions made in this tutorial¶
- Prior to working through this tutorial we recommend running through the TopiaryExplorer Overview Tutorial which will show you how to work with the basic features of TopiaryExplorer. This tutorial assumes that you already know how to load the TopiaryExplorer application and that you have some familiarity with the basic interface.
- Users must have a QIIME 1.3.0 install to complete the filtering steps.
Step 0. Compile your tree and sequences and verify format¶
To apply this process you’ll need your tree of interest in newick format, as well as the unaligned sequences represented by the tips in the tree in fasta format. The full tip identifiers must be the first space-separated field of the sequence identifiers in the fasta file. For example, if a record in your fasta file looks like:
>sequence1 some comment about the sequence
AAACCCCCCCCCCCCCCCCCAAAAAAAAAAATTTTTTTTT
The tip representing this sequence in the tree must be sequence1.
Step 1. Filter input tree to 99% identity¶
Assuming your input sequence collection is called inseqs_full.fna, run the following command to cluster into 99% OTUs:
pick_otus.py -m uclust -D -i inseqs_full.fna -s 0.99 -o otus
Step 2. Obtain the sequence identifier of each OTU centroid¶
Next we want to select the centroid of each OTU cluster as the tip we want to keep in the tree to represent the corresponding cluster of 99% identical sequences:
awk 'BEGIN {FS="\t"};{print $2}' otus/inseqs_full_otus.txt > tips_to_keep.txt
Step 3. Filter the phylogenetic tree¶
Finally we’ll filter the full tree to contain only the representative tips for each of the 99% OTUs:
filter_tree.py -i full.tre -o filtered_99.tre -t tips_to_keep.txt
Step 4. Create a new project in TopiaryExplorer¶
Open TopiaryExplorer and create a new project using the new project dialog. Open the tree and any metadata you have about the tips.
Step 5. Color the tree¶
Color the branches by a metadata field of interest. In this example we’re coloring by CATEGORY1.
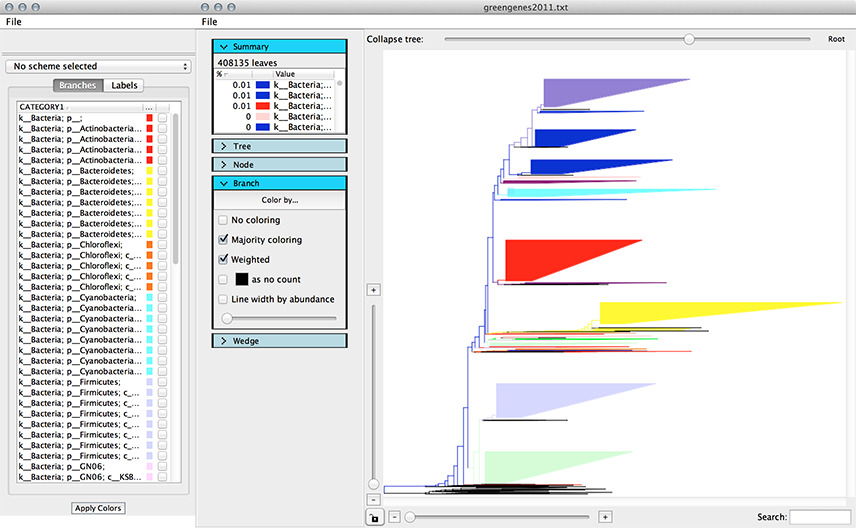
Note
Using the interpolate colors function with large trees can take a while (this example took a few minutes to color).